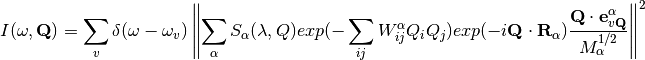Inelastic x-ray (IXS) and neutron (INS) scattering intensity maps:¶
The script make_TDS_DispersionIntensityCurves calculates inelastic scattering intensities for x-rays and neutrons. It has interactive capabilities to let you choose two modes at a given q-point and look for translations into other Brillouin zones which optimize the contrast (see below Interactions) It requires that you have performed the following operations beforehand:
- symmetrisation
- Fourier Transform
- Debye waller calculation for the temperature of choice
The usage is
make_TDS_DispersionIntensityCurves castep_filename input_filenamethe file castep_filename can be either the name of the original castep output the associated hdf5 file. In any case the associated hdf5 file must exist already (make_TDS_Simmetrization must be run beforehand)
The input_file must set the variables :
- Mandatories
- APPLYTIMEREVERSAL
- redStarts
- redEnds
- Nqlines
- Nfour_interp
- Temperature
- resolutionfile
- Saturation
- lowerLimit
- bottom_meV
- tth_max (degrees)
- If NEUTRONCALC==0 ( default )
- Lambda
- If NEUTRONCALC==1
- CohB
- NeutronE
- Optional
- branchWeight
- Eigscal
- UniqueIon
EXTRA OUTPUT
File summedoverQs.dat
contains two columns : energy , intensity. Obtained after summing over Qs
File alongtheline_TDS.dat
energy integrated intensity (TDS) for each q-value
INTERACTIONS
To toggle back and forth the Choosing mode (selection mode) type C with the graph as active window.
Choosing branch A :
when you are in selection mode left-click over the branch. The q-point also is selected according to the click position.
Choosing branch B :
right click
- To change manually selected points
: type mthis is sometimes necessary to disambiguate overlapping branches. This interaction opens an emacs editor with variable assignation. When you save and exit, the variables are updated with new values if you have modified them. You can, in this way, change the branch number and see if there are ambiguous neighbours.
- To run the optimisation
: type MThis launches a calculation of equivalent Qpoints in a wide range of cells (from -6 to 6 along all three axis ) which is centred over the initial q-point. New points are considered if they are physical : K < 2pi/lambda. The calculation writes the intensity of the two branches on file choosen_modes.txt.
The first three columns are the reduced reciprocal shift applied to the original Q point. The fourth is scattering angle theta in degree. The last three columns are Ia, Ib and Ia/Ib ( the constrast ) The lines are ordered in descending order of contrast.
The implemented formula is the following
where
is the branch index,
the atom index,
is the scattering factor for a given wavelenght ang angle,
are the Deby-Waller coefficients for the given temperature,
the atomic positions,
the atomic masses and
the eigenvectors.
The input variables are documented with docstrings below
-
make_TDS_DispersionIntensityCurves.APPLYTIMEREVERSAL= 1 this is one by default. Must be coherent with previous steps
-
make_TDS_DispersionIntensityCurves.redStarts= [[-2.0, -2.0, 0.0], [2.0, 2.0, 0.0]] The list of Q points at which calculation is made is done of N segments. The redStarts variable contains a list of N starting points in reciprocal space reduced units. Each segment i starts at redStarts[i] and ends at redEnds[i]. Within each segment the spacing between points is the segment lenght divided by Nqlines[i]
-
make_TDS_DispersionIntensityCurves.redEnds= [[2.0, 2.0, 0.0], [2.0, 2.0, 1.0]] see redStarts
-
make_TDS_DispersionIntensityCurves.Nqlines= [200, 100] see redStarts
-
make_TDS_DispersionIntensityCurves.Nfour_interp= 5 The number of points in each direction of the 3D reciprocal grid. The hdf5 file must contain a previous treatement done with the same parameter.
-
make_TDS_DispersionIntensityCurves.Temperature= 100 - The hdf5 file must include DW factors calculated ALSO at this temperature.
- The temperature at which DW factors have been calculated.
- Units are Kelvin
-
make_TDS_DispersionIntensityCurves.resolutionfile= '' a two column file : first the energy in cm-1, second the value of resolution function
-
make_TDS_DispersionIntensityCurves.Lambda= 1.0 For X-ray scattering : the wavelenght in Angstroems.
-
make_TDS_DispersionIntensityCurves.Saturation= 0 To limit intensity at peaks : intensity is saturated at this value
-
make_TDS_DispersionIntensityCurves.lowerLimit= 0 To correct the dynamical range, when displaying, the intensity is clipped to prevent it going below this value
-
make_TDS_DispersionIntensityCurves.bottom_meV= 0 Calculated eigenvalues are clipped to this value before use.
-
make_TDS_DispersionIntensityCurves.NEUTRONCALC= 0 This activate neutron scattering calculations.
-
make_TDS_DispersionIntensityCurves.CohB= None When neutroncalc is on, this must be a dictionary : for each atom name the Coherent lenght.
-
make_TDS_DispersionIntensityCurves.NeutronE= 0 When neutroncalc is on, the neutron energy in meV
-
make_TDS_DispersionIntensityCurves.branchWeight= None a list of weights : one per branch.
-
make_TDS_DispersionIntensityCurves.Eigscal= 0 EigScal==1 Intensity are calculated WITH only eigenvector scalar products ; ==2 Intensity are calculated WITH only eigenvector scalar products PLUS Mass factor
-
make_TDS_DispersionIntensityCurves.UniqueIon= -1 If >=0. Selects one ion. All the other will be silent.