tds2el: visualisations and fits¶
the following documentation has been generated automatically from the docstrings found in the source code.
running the code¶
tds2el is always runned with one arguments
tds2el inputfile
the inputfile contains driving options. Let’s call this file input.par. here an example
conf_file = "tds2el.par" filter_file = "filter.edf" images_prefix = "G19K45/set1_0001p_0" images_prefix = ".cbf" ascii_generators = "1~,2(z),2(y)" normalisation_file = "normalisation.dat"
conf_file: is the name of the file containings all the experimental parameters, and the parameters concerning your calculation options. You can compose it starting from the input file that you have used for the alignement, with updated refined parameters, and then adding some more options that we document here below.
filter_file : is the mask : it is one for good pixel and zero for pixel to be discarded. There is also another mechanism which discards data from the fit : when the data pixel has a negative value in the experimental image that’s a signal for the pixdel to be discarded.
images_prefix : the list of files will be built by the following lines of code (tds2el.py)
flist = glob.glob(params.images_prefix+"*"+ params.images_postfix) flist.sort()it means that the file ordering is given by the name ordering. This is an important point because on this depends the rotation angle given at each file thourgh the incremental step
paramsExp.phi += angular_stepascii_generators is used to determine the structure of elastic constants tensor.
normalisation_file contains two columns : filename ( with path) and denominator value. Look at the dedicated section of the documentation to see how you can generate one.
parallel or not parallel¶
when an harvesting is done, a huge amount of files needs to be readed. tds2el can be runned in parallel in this case
mpirun -n 8 tds2el input.parin the case you want to use 8 processors.
when the harvesting is already done, and you run fits and interpolations, the program does not use mpi.
It uses instead opencl for certain parts of the calculations but you always run the code simply by
tds2el inputfile
Working in the neighboroods of Bragg peaks¶
You can do :
- fine interpolations around selected bragg peaks
- optimise elastic constants to fit the neighbourood os selected Bragg peak
- The fit is done on the data. Not on the interpolation
The preliminary step for both the above operations consists in harvesting from the raw data, the vicinity of selected spots
preparation of the input file for harvesting¶
For the conf_file you can compose it starting from the input file that you have used for the alignement, with updated refined parameters, and then adding following options
DeltaQ_max = 0.15 DeltaQ_min = 0.04 load_from_file= None save_to_file = "harvest_spots.h5" DO_ELASTIC=1 threshold = 500000.0 if False: collectSpots = [[0,0,-12],[0,1,-8],[2,0,-8],[-4 , 4, 8 ],[-1,0,4],[-1,1,8],[-2,-1,-2],[2,0,-2] ] else: file_maxima = "tagged_selectedpeaks.p" REPLICA_COLLECTION = 0 # default to 0What do these options stand for ? :
DO_ELASTIC=1 activates enquiries concerning data points around Bragg peaks. ( the other less documented options DO_ELASTIC=0 is used to do rough harvesting of the volume for whole volume visualisation, but is less documented here and at the bottom)
DeltaQ_max, DeltaQ_min these numbers determine which area, around each (selected) peak, is harvested. These values are multiplied by the lenght of the longest reciprocal lattice vector ( {\bf a^* ,b^*, c^* } and thus converted to maximal radius and minimal radius. All the data points that fall within these limits are harvested.
load_from_file= None This option activates harvesting. It indicates that there is no previous knowledge from previous harvesting.
save_to_file = “harvest_spots.h5” The file the harvest will be written to.
collectSpots =[ points ] the spots around which data are collected. If alternatively a tagged maxima file_maxima is given ( tagged aftern alignement refinement) then the list will be formed with the hkl tags of the accepted spots. Note that a same hkl can give multiple peaks due to sweeping. At this version of tds2el hkl will be accepted only if all the spots tagged hkl are accepted. When a scan produces separated maxima for the same hkl the two blobs are fitted separately, this means that in future development of the code one could select just the selected ones in case of multiple hkl
REPLICA_COLLECTION : where it is set to 1 ( or True) then the collected sport are, besides those specified by collectSpots or maxima file, all those that can be obtained by symmetry operations.
threshold : when a pixel value exceed threshold no pixel values from that image are fitted , however the points of a blob exceeding threshold are used to calculate the position of the bragg peak in reciprocal space. This is done doing a weighted average. The averaging is not an exact procedure because of the pixel saturation but we hope that the saturated points are closely packed around the exact position. For centering be effective you have to harvest twice. The second you you use the option centering_file.
REMEMBER that harvesting can be parallel ( see above introduction)
TO enquire which hkl have been collected in a given and how many points each add the following ad rerun
load_from_file = "harvest_spots.h5" show_hkls = True
Recentering¶
If you want to obtain correct harvested Q, which are centered over bragg spot, calculated on the basis of the most intese pixels, you have to run the harvesting procedure twice. the second time you set centering_file to the previous harvest file
centering_file ="harvest_spots.h5"and you get a new harvest where Q are not calculated from geometry only , but also recorrection based on bragg spot position.
Simulating the Thermal Diffused Scattering from Elastic Constants¶
preparation of the input file¶
The preliminary step consists in doing a dry run to discover the structure of the elastic tensor. At this stage you just specify some spots to be fitted so that tds2el knows that you are going to fit something
DO_ELASTIC=1 load_from_file= "harvest_spots.h5" save_to_file = None fitSpots = Truetds2el will stop, in this dry run, because we have not yet provided the elastic constants, but before finishing will print on the screen the structure of the elastic tensor that it finds by simmetry operations
GET ELASTIC SYMMETRY 1.000000 A11 | 1.000000 A12 | 1.000000 A13 | | | --------------------------------------------------------------------------------------- 1.000000 A12 | 1.000000 A22 | 1.000000 A23 | | | --------------------------------------------------------------------------------------- 1.000000 A13 | 1.000000 A23 | 1.000000 A33 | | | --------------------------------------------------------------------------------------- | | | 1.000000 A44 | | --------------------------------------------------------------------------------------- | | | | 1.000000 A55 | --------------------------------------------------------------------------------------- | | | | | 1.000000 A66And the program stops if CGUESS is not provided. CGUEES can now be created with initial values, minima, maxima for A11,A12 and all the elastic constants that have been just printed in the dry run. Mind that the obtained structure could be considerably more complicated if the aAA,aBB.... differ more than a given tolerance from the nominal values. Correct them in this case, or alternatively change the tolerance which is so far hard-coded in tds2el.py
if abs(trace_matrici[ i*6+j ])>1.0e-6:IMPORTANT NOTICE : the alignement convention is the one of GIACOVAZZO where one align {\bf a^*} instead of {\bf a} along the X axis. For this reason some elastic constant may have different indexes than usual : A14 may become -A15 and so on.
To calculate the simulated diffused scattering you have to enter the values of elastic constants. You do this by setting the dictionary CGUESS. Here an example to get a simulation.
########################### # # Temperature and density # just in case # # At room temperature you can simultaneously scale # frequencies and intensity because of the bose factor getting linear in T. # Our fit is in this case insensitive to absolute scale # because the intensity factors are absorbed by # the fitting procedure. This because we dont have an absolute # scale of the absolute intensity : we dont know the exact shape # of the sample, beam profile, and many other factors Temperature = 300.0 # Kelvin 330 is the default value density = 2.71 # g/cc 2.71 is the default value ### DO_ELASTIC=1 load_from_file= "harvest_spots.h5" save_to_file = None fitSpots = True CGUESS={"A11": [ 254.307231832 , -1000 , 10000] , "A22": [ 254.307231832 , -1000 , 10000] , "A33": [ 144.467519525 , -1000 , 10000] , "A44": [ 120.580683913 , -1000 , 10000] , "A55": [ 169.691768154 , -1000 , 10000] , "A66": [ 169.691768154 , -1000 , 10000] , "A12": [ -7.53771283211 , -1000 , 10000] , "A13": [ 46.561752961 , -1000 , 10000], "A23": [ 46.561752961 , -1000 , 10000] } doMinimisation=0 visSim=1 testSpot_beta = 0.03 testSpot_Niter = 100 testSpot_N = 128 if True: # to fit just one spot collectSpots = [[-2,-2,1] ] else: file_maxima = "tagged_newpeaks.p" # or all the spots, # The visualisation is not so optimised as the fit of elastic constants # and runs on CPUs
CGUESS is the dictionary with initial values, minimum, maximum
doMinimisation=0 desactivate the optimisation ( the Fit to the data ).
REPLICA=0 if 1 collectSpots will be replicated by simmetry.
visSim=1 activates the visualisation ( if you do a simulation it is for looking at it ) which activates the interpolation around all the fitted spots. Visualisation is not so optimised, while fitting of the TDS is very oiptimised and runs on GPU. You can therefore when you set do_Minimisation=0, desactivate visSim, for the fit, outherwise you may have to wait some minutes more after the fit waiting for the visualisation to be calculated and written, and then rerun without minimisation, with visSim=1 and on a reduced set of spots.
How interpolation is done when activated by visSim=1: we use a formula which works both for the oversampled and under-sampled cases. We find the minimum of an object function which minimised a fidelity term, given by the squared norm of the difference between the data {bf y} , and the interpolated data, obtained interpolation the solution {bf x} given on the cubic grid, plus a regularisation term, multiplied by \beta, which is given by the squared norm of the gradient. The solution {bf x} is expressed mathematically as :
{\bf x^{*}} = argmin_{\bf x} \left( \frac{1}{2} \left\| Interp({\bf x}) - {\bf y} \right\|^2 + \beta \left\| \nabla {\bf x} \right\|^2 \right)
where \beta is given by the parameter testSpot_beta and the optimisation is done through testSpot_Niter iteration of FISTA algorithm. Full convergence is not needed, we are just doing visualisation: choosing a low number of iteration allows highlight the data granularity when data are more sparse then interpolation points. The relevant parameters are testSpot_beta testSpot_Niter.
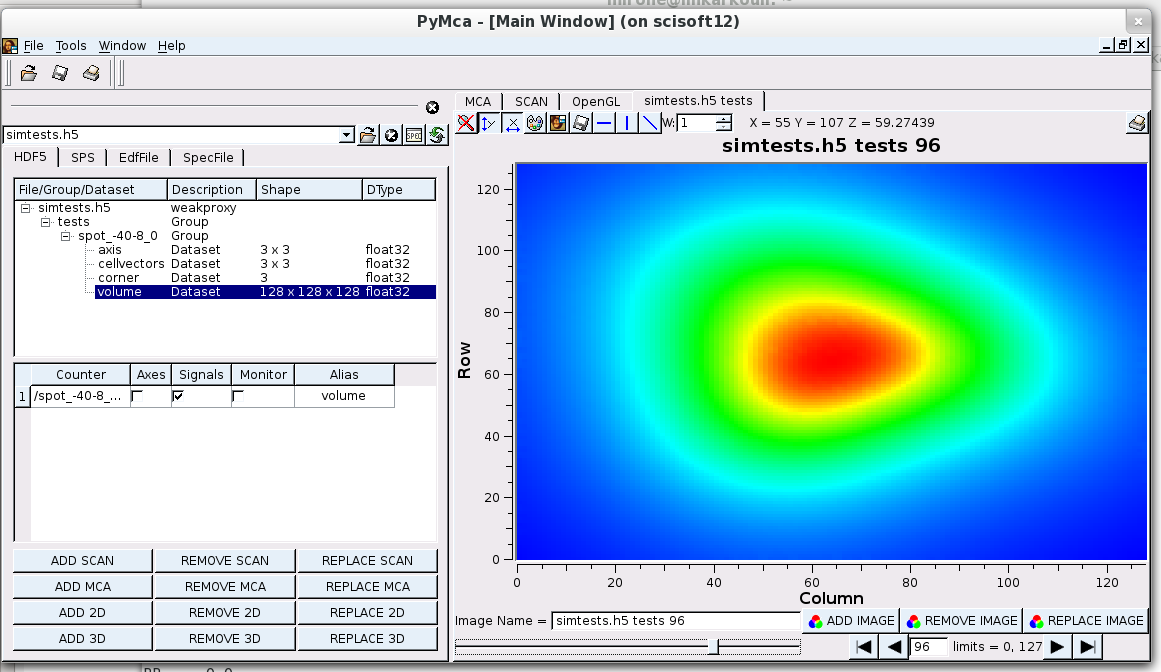
How you can look at the obtained result
pymca -f simtests.h5PERFORMANCE ISSUE for VISUALISATION: tds2el calculates simulated data at all the data harvested data points(which are replicated by simmetry). Then for each one of them, an iterative interpolation is done for the cubic grid. It is this last operation that takes time. Therefore you might :
- select a not too long list of spots
DOING THE FIT¶
preparation of the input file¶
The input file is basically the same as for simulating the spots but with doMinimisation set to True and, if you dont want to wait for subsequent interpolations, visSim=0. The obtained parameters will be printed at the screen once the fit has converged. They are also written on file parametri.txt
DO_ELASTIC=1 load_from_file= "harvest_spots.h5" save_to_file = None collectSpots = [[-2,-1,-2],[-1,0,4],[0,1,-8] ,[2,0,-2], [-2,-1,-2], ] CGUESS={"A11": [ 254.307231832 , -1000 , 10000] , "A33": [ 144.467519525 , -1000 , 10000] , "A44": [ 120.580683913 , -1000 , 10000] , "A66": [ 169.691768154 , -1000 , 10000] , "A13": [ -7.53771283211 , -1000 , 10000] , "A15": [ 46.561752961 , -1000 , 10000] } doMinimisation=1 visSim=0PERFORMANCE ISSUE : very important
you are going to do a lot of tests before producing an useful result. On the other hand the data amount is huge. TO go really fast use, in your conf_file
nred=10or higher. One point over nred only will be used. This might eventually accelerate also the interpolation because you have less data points, but in this case you will have more coarse granularity and you might need a higher beta. If you dont set nred it is nred=1 by default.
ANOTHER PERFORMANCE ISSUE :
The simulation calculation are done with openCL, by default on GPU. On a non GPU machine use
devicetype="CPU"to use all available cores.
Convergenge control :
ftols is by default=0.00001. If you set ftol=0.001, in conf_file, convergence condition will be satisfied sooner.
Fine retuning of angles r1 r2 r3 and cell parameters a,b,c¶
THIS PART IS NOT SO USEFUL IF YOU ARE RECENTERING (Option centering_file =”harvest_spots.h5”). The fit is done not only on the elastic constants ( and a pro-spot factor plus baseline ) but also on Alignement parameters r1 r2 r3 and cell parameters a,b,c. ( For more informations about alignement geometry See thesis of Mathias MEYER Construction of a multi-purpose X-ray CCD detector and its implementation on a 4-circle kappa goniometer) Université de LausanneAs default these Alignement corrections are constrained between 0- and 0+
Their value is given by the internal A_variables symbol whose default is
A_variables = [ minimiser.Variable(0.0,0.0,0.0) for i in range(6+4+2+2) ] # (6+4+2+2) parameters corresponding to # r1,r2,r3, AA, BB, CC , # omega, alpha, beta, kappa # d1,d2, # det_origin_X, det_origin_YThese are variation variables which are added to their corresponding parameter. You can free (or not) the optimisation for these variables by opening (or closing) their min-max interval
A_variables = ( [ minimiser.Variable(0.0,-1.0e-1,1.0e-1) for i in range(3) ] + [ minimiser.Variable(0.0,-3.0e-2,3.0e-2) for i in range(3) ] + [ minimiser.Variable(0.0,0.0,0.0) for i in range(8) ] )You can add the above lines to the conf_file file The optimised value is the correction ( 0 means no correction) and the final value is printed at the end of the fit, on screen and also in file parametri.txt The following lines allow to open or close selected group of intervals with less hand editing
activ =1*0 activ1=1*0 activ2=1*0 activ3=1*0 A_variables = ( [ minimiser.Variable(0.0,-1.5*activ1 ,1.5*activ1 ) for i in range(1) ]+ # r1, [ minimiser.Variable(0.0,-1.5*activ1 ,1.5*activ1 ) for i in range(1) ]+ # r2 [ minimiser.Variable(0.0,-1.5*activ1 ,1.5*activ1 ) for i in range(1) ]+ # r3, [ minimiser.Variable(0.0,-0.1*activ2 ,0.1*activ2 ) ,-1, minimiser.Variable(0.0,-0.1*activ2 ,0.1*activ2 ) ]+ # AA, BB, CC [ minimiser.Variable(0.0,-2.0*activ3 ,2.0*activ ) , minimiser.Variable(0.0,-2.0*activ3,2.0*activ3) , minimiser.Variable(0.0,-2.0*activ3,2.0*activ3) , minimiser.Variable(0.0,-2.0*activ3,2.0*activ3) , ]+ # omega, alpha, beta, kappa [ minimiser.Variable(0.0,-0.2*activ,+0.2*activ) for i in range(2) ] + # d1,d2, [ minimiser.Variable(0.0,-5*activ,+5*activ) for i in range(2) ] # det_origin_X, det_origin_Y )