tds2el_extract_maxima: Extracting positions and screening of the spots¶
the following documentation has been generated automatically from the docstrings found in the source code.
tds2el_extract_maxima is always runned with two arguments
tds2el_extract_maxima input_file
The maxima search operation profits from parallelism
mpirun -n 12 tds2el_extract_maxima input_file
Three different kind of task can be activated by setting to True the corresponding switch parameter
- By default the 3D peaks are searched into a stack of images
- drawmask = True. This option sums up a small numer of images of the stack. It displays them and a mask can be drawn. The mask is then written to a file and can be used during the peak search to set to zero anomalous pixels(filter_file option).
- review = True. Visual navigation in the found maxima. They can be tagged as good or bad. In the future the possibility of introducing a mask-per-blob could be considered. But it is maybe better to do it in the reciprocal space.
example de fichier d’ input
images_prefix = "G19K45/set1_0001p_" # Let the following images_prefix_alt to None and dont set it ( default None) # It is used for reviewing with comparison of same position locations # versus an alternative dataset # images_prefix_alt = "G19K45/set1_0001p_" images_postfix = ".cbf" ## peaks are searched, having a radius peak_size # and with maximum > threshold_abs and > image_max*threshold_rel threshold_abs=40000 threshold_rel=0.00 peak_size = 2 spot_view_size = 10 # Filtering, OPTIONAL or set it to None for no mask filtering of bad pixels # filter_file = None filter_file = "filter.edf" ## medianize can be 1 or 0 ( True or False) ## The peak is always searched into a stack ## of spot_view_size + 1 + spot_view_size slices ## When the option is activated each slice will be beforehand ## subtracted with the median of the slices going from the spot_view_size^th preceeding ## to the spot_view_size^th following. Spot_view_size should be greaer than peak_size medianize = 1 ## Let this advanced option as it is for now, or dive into the code. ## The rationale here would be that in the first step on works ## locally on spot_view_size + 1 + spot_view_size slices, ## in the second step one can work globally on the found peaks globalize = (0,0) ## The file where found peaks are written ## or, when reviewing, read. file_maxima = "maxima.p" ### if yes, a graphical window allows for the selection of the mask ### Closing the window will propose to write on newmask.edf drawmask = False ## Allows to have a simultaneous and animated view of the blobs. ## Animated thumnails. ## They can be selected of deselected. When clicking on the ## thumbnails a larger 3D explorer is charged with ## a (review_size+1+review_size )^3 stack review = False review_size = 50
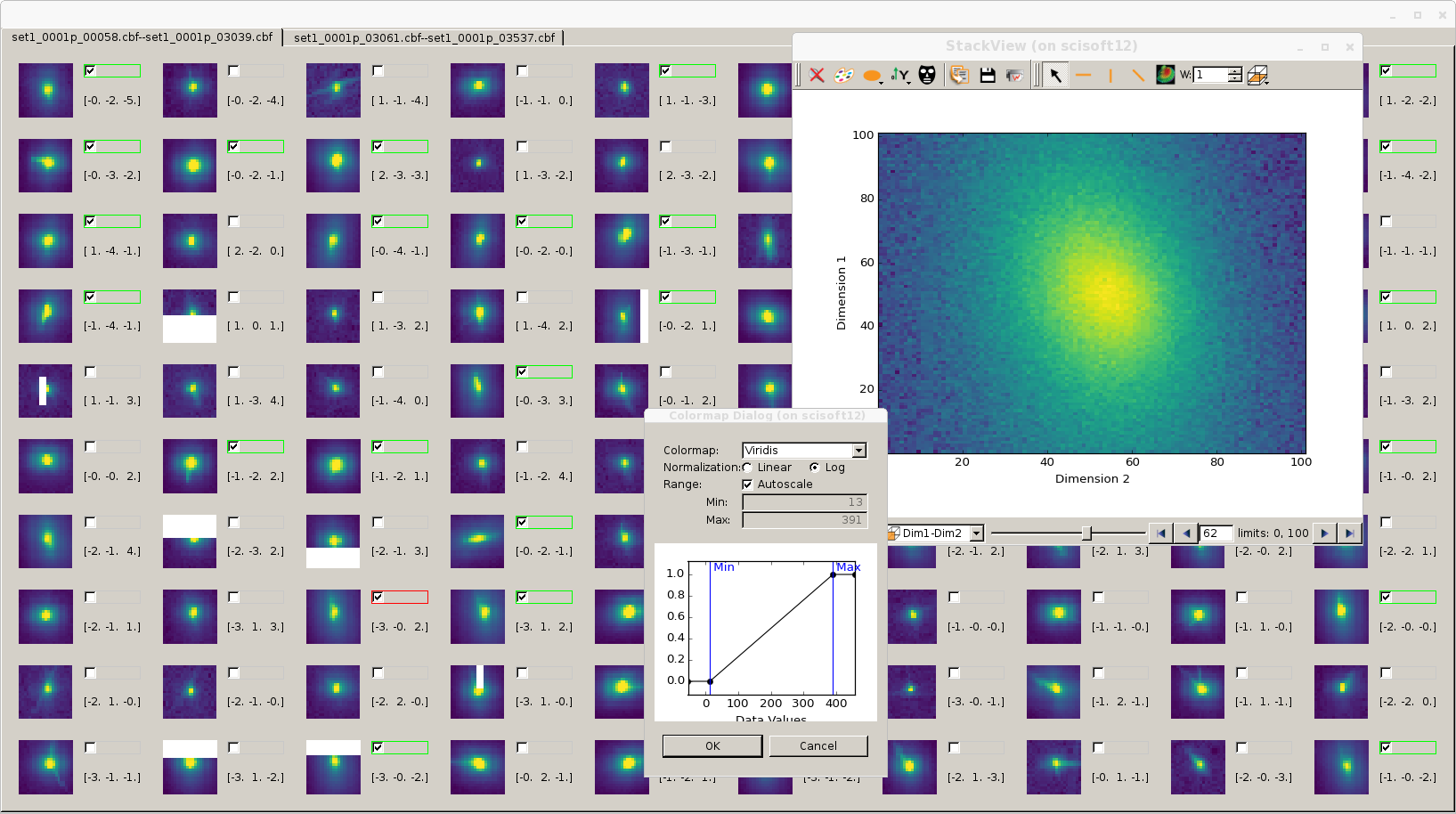
The following image illustrates the use of the viewer. Peaks can be selected and deselected. When closing the windows you are asked if yes or not you want to save the maxima on a new file. The file can be reused for the further steps: geometry alignement and elastic constants fitting. The alignements steps create a new file where peaks are tagged by hkl. The image below is the run of the viewer on such a file, hkl is displayed. Each icon is animated movie sweeping the small (2*spot_view_size+1)**3 cube around the peak in image space: each frame correspond to an experimental image. If you let the mouse pointer idle on an icon a toolitip appear mentioning further metadata od the spot. If you click the 3D stack viewer from Silx is charged with a larger (2*review_size+1)**3 data volume of the data. Whe you have loaded an image and working on it, its checkbox is red-framed to remember what you are working on. The alt-data if any will be displayed as an additional companion icon on the right of the checkbox
The thumbnails are generated in extraction phase using a Silx widget. At present the widget needs to have acces to a graphical display and interact with it, this might be a performance issue if the extraction is runned remotely in some cases of slow network. With NX ( no-machine) the performances are good even remotely.
-
class
extract_maxima2.MyStack(parent=None, resetzoom=True, backend=None, autoScale=False, logScale=False, grid=False, colormap=True, aspectRatio=True, yinverted=True, copy=True, save=True, print_=True, control=False, position=None, mask=True)[source] -
closeEvent(self, QCloseEvent)[source]
-
-
class
extract_maxima2.PeakoScope(maxima_data, review_size, flist, flist_alt, parent=None)[source] -
closeEvent(self, QCloseEvent)[source]
-