Creating Normalisation Factors¶
Normalisation may be important when absolute values need to be found, because in this case comparisons are done between two temperatures or two other parameters, for the same sample, measured at two different times.
Otherwise if the intensity normalisation vary slowly, and you search relative constants, it may not be a problem when variation are small during the sweeping of a single spot. (Tds2el fits separately the same hkl which has been swept two or more times during a scan).
When normalisation is important, we need a two columns file : first column the name of the file; second column a number. Rescale th is number not far from 1 so that you dont loose the idea of the detector signal during the following analysis phases.
This file must be provided by you because metadata format, if any, is different from setup to setup. Sometimes one uses a measurement, sometimes the intensity of the beam, sometimes nothing at all. When something is written each one uses a different name.
We are going to produce such a file based on a time indication that we found in the files header. If you dont have even that indication, than you better preserve the file time-stamp when copying or tranfert data and use that.
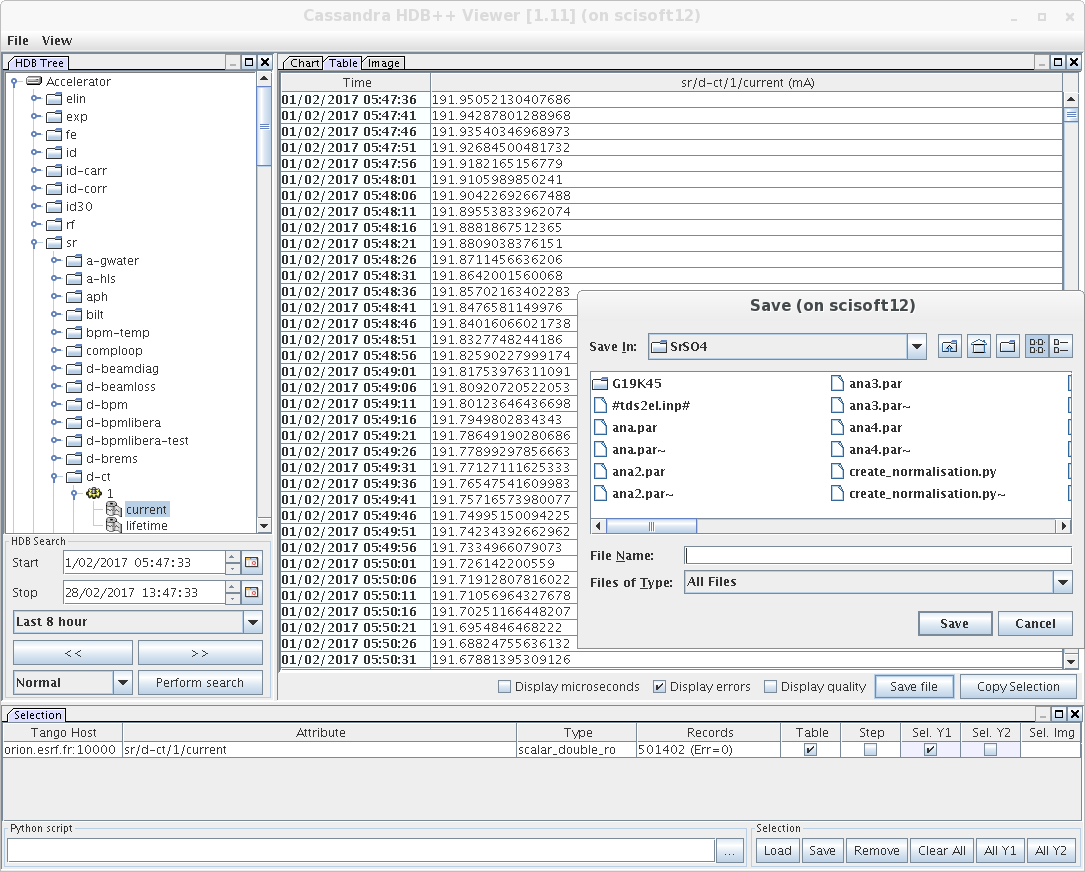
We are going to use beam intensity of the Esrf machine, that we can retrieve on rnice8 by using hdb_query
/segfs/tango/release/java/scripts/hdb_query
and then select the proper informations
You select Accelerator->sr->c-ct->1->current, then you ask a table, choose a period start-end that covers your experiment, click on perform Search, you display the table tab and finally you save on a file, let’s call it INTENSITY.dat. The produced files is like this
# File generated from hdbviewer application
#
HDB Date HDB Time sr/d-ct/1/current (mA)
01/02/2017 05:47:36 191.95052130407686
01/02/2017 05:47:41 191.94287801288968
01/02/2017 05:47:46 191.93540346968973
01/02/2017 05:47:51 191.92684500481732
01/02/2017 05:47:56 191.9182165156779
01/02/2017 05:48:01 191.9105989850241
01/02/2017 ....... ..............
...............................
.................
Now we are going to produce the desired two columns file with a python script ( should be easily adaptable to your case). We rescale by 170 because the experiment time lapse corresponded more or less to this value
import datetime
import fabio
import glob
from scipy.interpolate import interp1d
import time
import string
def get_timeintensity_interpolator(filename):
times = []
ints = []
s=open(filename,"r").read()
if len(s)<200:
return string.atof(s)
sl = string.split(s,"\n")
for l in sl[3:]:
if "Err" in l or "Advi" in l:
continue
l=string.split(l)
if len(l)==0: break
day,month, year = string.split(l[0],"/")
hours,mins,secs = string.split(l[1],":")
dt = datetime.datetime(int(year), int(month), int(day), int(hours), int(mins), int(secs))
s = time.mktime(dt.timetuple())
# print s, float( l[2])
times.append(s)
ints.append(float( l[2] ))
return interp1d(times, ints)
interpolator = get_timeintensity_interpolator("INTENSITY.dat")
output = open("normalisation.txt","w")
fl = glob.glob("G19K45/set*cbf")
for fn in fl:
print fn
im = fabio.open( fn,"r")
tempo = im.getheader()["_array_data.header_contents"].split("\n")[1][2:]
year, month,day,hours,mins,secs = tempo[:4], tempo[5:7], tempo[8:10],tempo[11:13], tempo[14:16], tempo[17:]
dt = time.mktime( datetime.datetime(int(year), int(month), int(day), int(hours), int(mins), int(float(secs))). timetuple() )
output.write("%s %e\n"%(fn, interpolator(dt)/170.0) )
and you get the firl normalisation.txt
G19K45/set1_0001p_02196.cbf 1.003309e+00
G19K45/set1_0001p_03329.cbf 1.003275e+00
G19K45/set1_0001p_00697.cbf 1.003347e+00
G19K45/set1_0001p_03052.cbf 1.003279e+00
G19K45/set1_0001p_01294.cbf 1.003335e+00
G19K45/set1_0001p_02050.cbf 1.003315e+00
G19K45/set1_0001p_00294.cbf 1.003358e+00
G19K45/set1_0001p_00479.cbf 1.003355e+00
G19K45/set1_0001p_00642.cbf 1.003351e+00
G19K45/set1_0001p_01609.cbf 1.003325e+00
G19K45/set1_0001p_01924.cbf 1.003320e+00
G19K45/set1_0001p_01853.cbf 1.003320e+00
G19K45/set1_0001p_02946.cbf 1.003283e+00
G19K45/set1_0001p_02220.cbf 1.003309e+00
..........................................